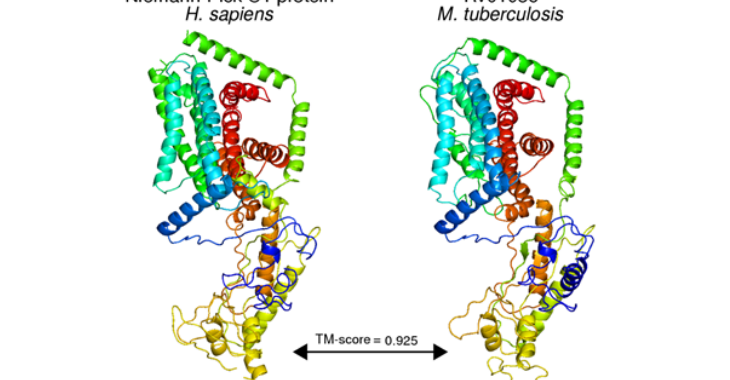
In this work, we developed protocols to (1) systematically curate discoveries from literature and (2) identify structural matches likely to share form and function with the gene product of interest. We employed these protocols on the primary reference strain (H37Rv) of Mycobacterium tuberculosis and annotated 713 (41.2%) of previously unannotated genes. These annotations uncovered dozens of putative host-interacting proteins, and dozens more of proteins performing functions implicated in basic metabolism, virulence, resistance, and tolerance. More broadly, this work provides an approach to functional annotation capable of deriving additional functional insight into well-studied genomes, and for high-throughput annotation of new and less-studied genomes