Slideshow
NEWS
-
Our recent publication “Drivers and sites of variability in the DNA adenine methylomes of 93 Mycobacterium tuberculosis complex clinical isolates”, published in eLife
-
SDSU TB Day 2020
-
Featured: SDSU Researcher Works to Combat Drug Resistance
-
Check out our latest pre-print, where we annotate hundreds of genes, cutting away at the M. tuberculosis “hypotheticome”
Laboratory for Pathogenesis of Clinical Drug Resistance and Persistence
Led by Dr. Faramarz Valafar, we are an NIH funded laboratory through the National Institute for Allergy and Infectious Diseases (NIAID).
We study the genetic (including dark genetic matter), epigenetic, metagenomic bases for treatment failure in infectious diseases. This includes study of evolutionary path and emergence of enhanced antibiotic resistance, virulence, fitness, and host pathogen interactions.
Our primary project focuses on tuberculosis (TB), which recently surpassed HIV/AIDS as the infectious disease causing the highest mortality. Over 10 million people develop the active form of the disease and 1.5 million people die from it annually. Antibiotic resistance in Mycobacterium tuberculosis (Mtb), the causative agent of TB, is a serious threat to the World Health Organization’s efforts in global TB control. In some parts of the world, 75% of all active TB cases are multi-drug resistant.
Our funding provides support for our research projects in San Diego and collaborations with seven international sites in Stockholm, Antwerp, Minsk, Cape Town, Mumbai, Riga, and Chisinau. In collaboration with hospitals and public health agencies in these cities, we study the emergence and mechanisms of antibiotic resistance, virulence, fitness, and the molecular basis for other clinically relevant phenotypes in Mtb.
Evolution of Drug Resistance
We use genomics to reconstruct how antibiotic resistant, hypervirulent, and persistent phenotypes emerge
Read More
Genomics & Epigenomics
We assemble SMRT-sequencing data into Mtb genomes and methylomes de novo and identify anomalies at single-base resolution
Read More
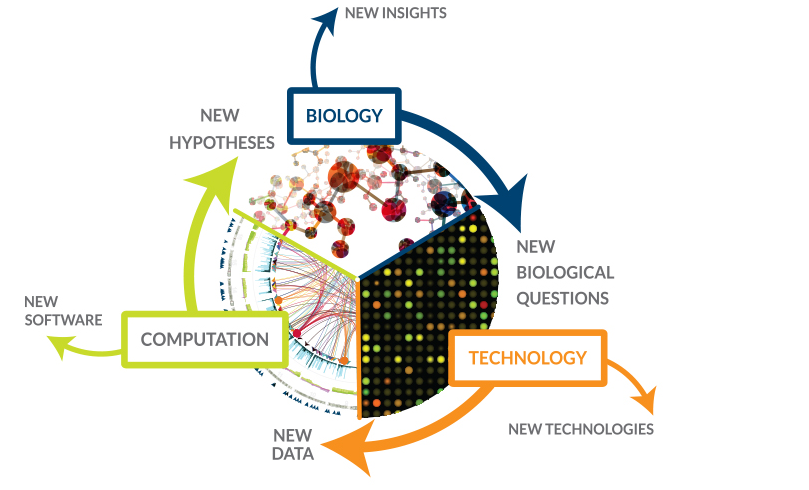
Systems Biology
We investigate how variations combine at the systems level to affect pathogenic virulent, and resistant phenotypes
Read More
Annotation & Functional Genomics
We develop and utilize tools to annotate how interesting variations or novel genetic material affect Mtb function.
Read MoreTechnology
Through single-molecule, real-time (SMRT) sequencing, we investigate the molecular basis and pathogenesis of clinically relevant phenotypes.
Read More